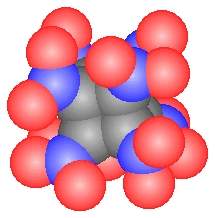
 US chemists have synthesized a new type of explosive that could be more powerful than the most potent non-nuclear compound known.
US chemists have synthesized a new type of explosive that could be more powerful than the most potent non-nuclear compound known.
The compound was synthesized by Philip Eaton and Mao-Xi Zhang at the University of Chicago by building on the building block hydrocarbon cubane. Cubane, first synthesized by Eaton and Thomas Cole, in 1964, is one of the most dense hydrocarbons ever made and has always been known as a highly strained compound that stores a great deal of energy,’ explains Eaton.
 Eaton and Zhang have developed a methodology involving non-standard procedures at each step to swap cubane’s corner hydrogen atoms with nitro groups – ultimately producing octanitrocubane. Nitro groups are common constituents of explosives for example in trinitrotoluene (TNT). The nitro groups provide oxygen for combustion of the carbon to carbon dioxide and release of nitrogen gas – explosively.
Eaton and Zhang have developed a methodology involving non-standard procedures at each step to swap cubane’s corner hydrogen atoms with nitro groups – ultimately producing octanitrocubane. Nitro groups are common constituents of explosives for example in trinitrotoluene (TNT). The nitro groups provide oxygen for combustion of the carbon to carbon dioxide and release of nitrogen gas – explosively.
Eaton says the compound is shock insensitive so he can safely hit it and its hepta precursor with a hammer. However, deliberately detonating it is highly explosive. Calculations show that the detonation velocities and pressures will be far greater than those of TNT and more explosive than HMX (high-melting explosive. Eaton says octanitrocubane may prove to be the most powerful non-nuclear explosive around.
Octanitrocubane might have applications in mining and demolition and in propellant applications. Angew Chem Int Ed Engl, 2000, 39, 401
Throwing out the silicon

‘Silicon chips with everything’ was one of the great puns of the 1970s and for more than thirty years the tiny etched elemental slivers have spread like a rash over every facet of our lives – find their place in running phones, TVs, washing machines, medical equipment, cars, and of course computers.
Getting more power out a silicon chip means cramming in smaller and smaller logic units into the same space. But there is a limit dictated both by the shortest wavelength X-rays we could use to ‘print’ the features and the quantum uncertainty that may arise as these features are packed closer and closer together. Nature, of course, may have provided scientists with a model answer.
Nucleic acids RNA and DNA – the carriers of one of the neatest digital information systems we know – might in the coming decades begin to displace the computers of doped silicon and its inorganic cousins for certain computational problems and replace it with chips based on the stuff of life. Leonard Adleman at the University of Southern California was the first scientist to begin using nucleic acids to solve mathematical puzzles. In 1994, he solved a version of the ‘traveling salesman’ problem. He used the DNA bases to represent combinations of stretches of the salesman’s journey and an enzyme that could recognize and cleave those that did not correspond to an optimum journey to compute the most efficient route. Molecular biology tools – enzymes, nucleic acids and such, were thus used to demonstrate the feasibility of carrying out computations at the molecular level.
Two papers published in Nature and the Proceedings of the National Academy of Sciences demonstrate the proof of principle that shows just how plausible a nucleic acid computer might ultimately be.
 Evolutionary biologist Laura Landweber collaborated with computer scientist Richard Lipton, Dirk Faulhammer and Anthony Cukras to see whether they could solve a basic chess problem. The so-called ‘knight problem’ asks how many and where can one place knights on a chessboard so they cannot attack each other. Such a problem is known mathematically as an example of an ‘NP-complete’, which means the possible answers increase exponentially with increasing variables – in this case squares on the chessboard. The knight problem represents a class of mathematical problems that can best be solved by simple brute-force computation rather than a complex logical approach. So, to number crunch the problem, the team built a computational system from 1024 different strands of ribonucleic acid (RNA) combinations to look into this problem. Each strand of RNA represents a configuration of pairs of knights and to simplify the demonstration the system is equivalent to a nine-square chess board (3 x 3) so that there are a ‘mere’ 512 possible combinations of knights rather than the millions possible with a standard 64-square board.
Evolutionary biologist Laura Landweber collaborated with computer scientist Richard Lipton, Dirk Faulhammer and Anthony Cukras to see whether they could solve a basic chess problem. The so-called ‘knight problem’ asks how many and where can one place knights on a chessboard so they cannot attack each other. Such a problem is known mathematically as an example of an ‘NP-complete’, which means the possible answers increase exponentially with increasing variables – in this case squares on the chessboard. The knight problem represents a class of mathematical problems that can best be solved by simple brute-force computation rather than a complex logical approach. So, to number crunch the problem, the team built a computational system from 1024 different strands of ribonucleic acid (RNA) combinations to look into this problem. Each strand of RNA represents a configuration of pairs of knights and to simplify the demonstration the system is equivalent to a nine-square chess board (3 x 3) so that there are a ‘mere’ 512 possible combinations of knights rather than the millions possible with a standard 64-square board.
The team used a targeted ribonuclease enzyme which seeks out specific combinations of bases along the strands of RNA and cleaves those that are marked by base-pairing to short DNA probes. By choosing to mark exactly the right bases the enzyme will cleave only those strands of RNA representing an incorrect combination – the wrong solutions to the knight problem, explains Landweber. All the team then needed to do was to analyze the remaining ‘whole’ RNA strands to find the answers to the problem.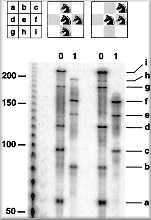
Landweber and her colleagues correctly identified 43 solutions to the knight problem with their RNA computer. It’s a start but is it a total solution? Landweber explains: ‘Each molecule represents a solution. We do not need to recover all of the actual solutions, since that would just be tedious, and that is not the challenge. By sampling, if we collected a large enough set we would probably get them all, keeping in mind that some would come up more than once just by the probability distribution.’ The team also points out that their system also kicked up one incorrect solution, which re-emphasizes the need for error checking within any computer system.
The main advantage, of course, adds Landweber, is that nucleic acids can have a much higher information density than any silicon chip – a gram of dried DNA could hold as much information as a trillion CD-ROMs, it is thought.
Meanwhile, a team at the University of Wisconsin-Madison has demonstrated that it might be possible to incorporate DNA itself into more solid devices that might ultimately be interconnected to electronic components. The free-floating world of solutions has always been a barrier to the acceptance of molecular computing. The main question is always how would a chemical solution be connected to real-world input and output devices such as keyboards and monitors?
 Chemist Lloyd Smith and his colleagues have now shown they can attach a potentially computing set of DNA strands by one end to a gold-coated glass surface. They use their DNA-device to eliminate incorrect solutions from another NP-complete problem known as the satisfiability problem, which is essentially the knight problem phrased differently. By exposing the tethered strands to free-floating strands carrying sequences complementary to those satisfying each criterion met by the correct solution they could produce double helices. The incorrect solutions at each step are then exposed to a cleaving enzyme. Gentle heating regenerates single strands and the next DNA complement is added as the next criterion and so on. Decoding the single remaining double strands provides the problems solution.
Chemist Lloyd Smith and his colleagues have now shown they can attach a potentially computing set of DNA strands by one end to a gold-coated glass surface. They use their DNA-device to eliminate incorrect solutions from another NP-complete problem known as the satisfiability problem, which is essentially the knight problem phrased differently. By exposing the tethered strands to free-floating strands carrying sequences complementary to those satisfying each criterion met by the correct solution they could produce double helices. The incorrect solutions at each step are then exposed to a cleaving enzyme. Gentle heating regenerates single strands and the next DNA complement is added as the next criterion and so on. Decoding the single remaining double strands provides the problems solution.
Smith concedes that nucleic acid computing is still a long way from approaching silicon technology. This is simply a test-bed for working out an improved and simpler chemistry for DNA computing, he says. Test-bed or not with the possibility of an information density a million million times higher than a CD-ROM there is an enormous incentive to get to grips with the shrinking world of computers.
L. M. Adleman et al., Science, 1994, 266, 5187.
L. Landweber et al., Proc. Natl. Acad. Sci. (USA), 2000, 97, 1385.
L.M. Smith et al., Nature, 2000, 403, 175.
Got an iPod? Sick of having to charge it up at the electric outlet? Then try maple syrup or cola!
 A couple of years ago NASA released satellite images of Santa Claus’ summer hideaways, turns out there are two locations in the USA where old Father Christmas likes to spend the lazy summer months. Santa Claus, Georgia (left) and Santa Claus, Indiana (right), are typical of Santa’s summer retreats, says NASA, offering Santa and Mrs Claus, all the reindeer and elves the usual amenities and a nice warm respite from their chilly Polar home during the off season.
A couple of years ago NASA released satellite images of Santa Claus’ summer hideaways, turns out there are two locations in the USA where old Father Christmas likes to spend the lazy summer months. Santa Claus, Georgia (left) and Santa Claus, Indiana (right), are typical of Santa’s summer retreats, says NASA, offering Santa and Mrs Claus, all the reindeer and elves the usual amenities and a nice warm respite from their chilly Polar home during the off season. Anyway, the official address to send your Letter to Santa is: Mr S Claus, North Pole, H0H OH 0, don’t worry if that zipcode looks a little odd, just try saying it out loud and you’ll feel okay.
Anyway, the official address to send your Letter to Santa is: Mr S Claus, North Pole, H0H OH 0, don’t worry if that zipcode looks a little odd, just try saying it out loud and you’ll feel okay. US chemists have synthesized a new type of explosive that could be more powerful than the most potent non-nuclear compound known.
US chemists have synthesized a new type of explosive that could be more powerful than the most potent non-nuclear compound known. Eaton and Zhang have developed a methodology involving non-standard procedures at each step to swap cubane’s corner hydrogen atoms with nitro groups – ultimately producing octanitrocubane. Nitro groups are common constituents of explosives for example in trinitrotoluene (TNT). The nitro groups provide oxygen for combustion of the carbon to carbon dioxide and release of nitrogen gas – explosively.
Eaton and Zhang have developed a methodology involving non-standard procedures at each step to swap cubane’s corner hydrogen atoms with nitro groups – ultimately producing octanitrocubane. Nitro groups are common constituents of explosives for example in trinitrotoluene (TNT). The nitro groups provide oxygen for combustion of the carbon to carbon dioxide and release of nitrogen gas – explosively.
 Evolutionary biologist Laura Landweber collaborated with computer scientist Richard Lipton, Dirk Faulhammer and Anthony Cukras to see whether they could solve a basic chess problem. The so-called ‘knight problem’ asks how many and where can one place knights on a chessboard so they cannot attack each other. Such a problem is known mathematically as an example of an ‘NP-complete’, which means the possible answers increase exponentially with increasing variables – in this case squares on the chessboard. The knight problem represents a class of mathematical problems that can best be solved by simple brute-force computation rather than a complex logical approach. So, to number crunch the problem, the team built a computational system from 1024 different strands of ribonucleic acid (RNA) combinations to look into this problem. Each strand of RNA represents a configuration of pairs of knights and to simplify the demonstration the system is equivalent to a nine-square chess board (3 x 3) so that there are a ‘mere’ 512 possible combinations of knights rather than the millions possible with a standard 64-square board.
Evolutionary biologist Laura Landweber collaborated with computer scientist Richard Lipton, Dirk Faulhammer and Anthony Cukras to see whether they could solve a basic chess problem. The so-called ‘knight problem’ asks how many and where can one place knights on a chessboard so they cannot attack each other. Such a problem is known mathematically as an example of an ‘NP-complete’, which means the possible answers increase exponentially with increasing variables – in this case squares on the chessboard. The knight problem represents a class of mathematical problems that can best be solved by simple brute-force computation rather than a complex logical approach. So, to number crunch the problem, the team built a computational system from 1024 different strands of ribonucleic acid (RNA) combinations to look into this problem. Each strand of RNA represents a configuration of pairs of knights and to simplify the demonstration the system is equivalent to a nine-square chess board (3 x 3) so that there are a ‘mere’ 512 possible combinations of knights rather than the millions possible with a standard 64-square board.
 Chemist Lloyd Smith and his colleagues have now shown they can attach a potentially computing set of DNA strands by one end to a gold-coated glass surface. They use their DNA-device to eliminate incorrect solutions from another NP-complete problem known as the satisfiability problem, which is essentially the knight problem phrased differently. By exposing the tethered strands to free-floating strands carrying sequences complementary to those satisfying each criterion met by the correct solution they could produce double helices. The incorrect solutions at each step are then exposed to a cleaving enzyme. Gentle heating regenerates single strands and the next DNA complement is added as the next criterion and so on. Decoding the single remaining double strands provides the problems solution.
Chemist Lloyd Smith and his colleagues have now shown they can attach a potentially computing set of DNA strands by one end to a gold-coated glass surface. They use their DNA-device to eliminate incorrect solutions from another NP-complete problem known as the satisfiability problem, which is essentially the knight problem phrased differently. By exposing the tethered strands to free-floating strands carrying sequences complementary to those satisfying each criterion met by the correct solution they could produce double helices. The incorrect solutions at each step are then exposed to a cleaving enzyme. Gentle heating regenerates single strands and the next DNA complement is added as the next criterion and so on. Decoding the single remaining double strands provides the problems solution.